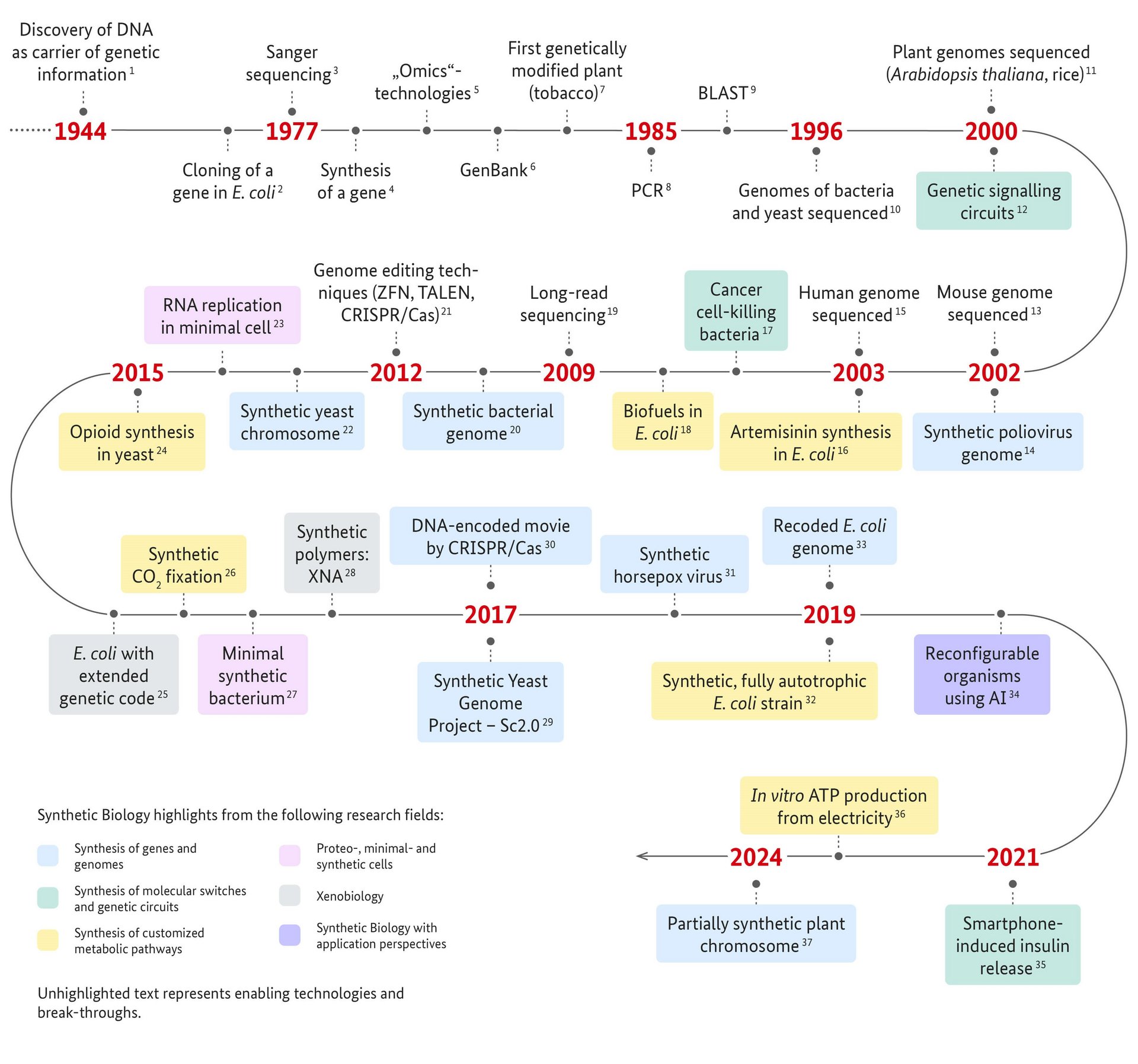
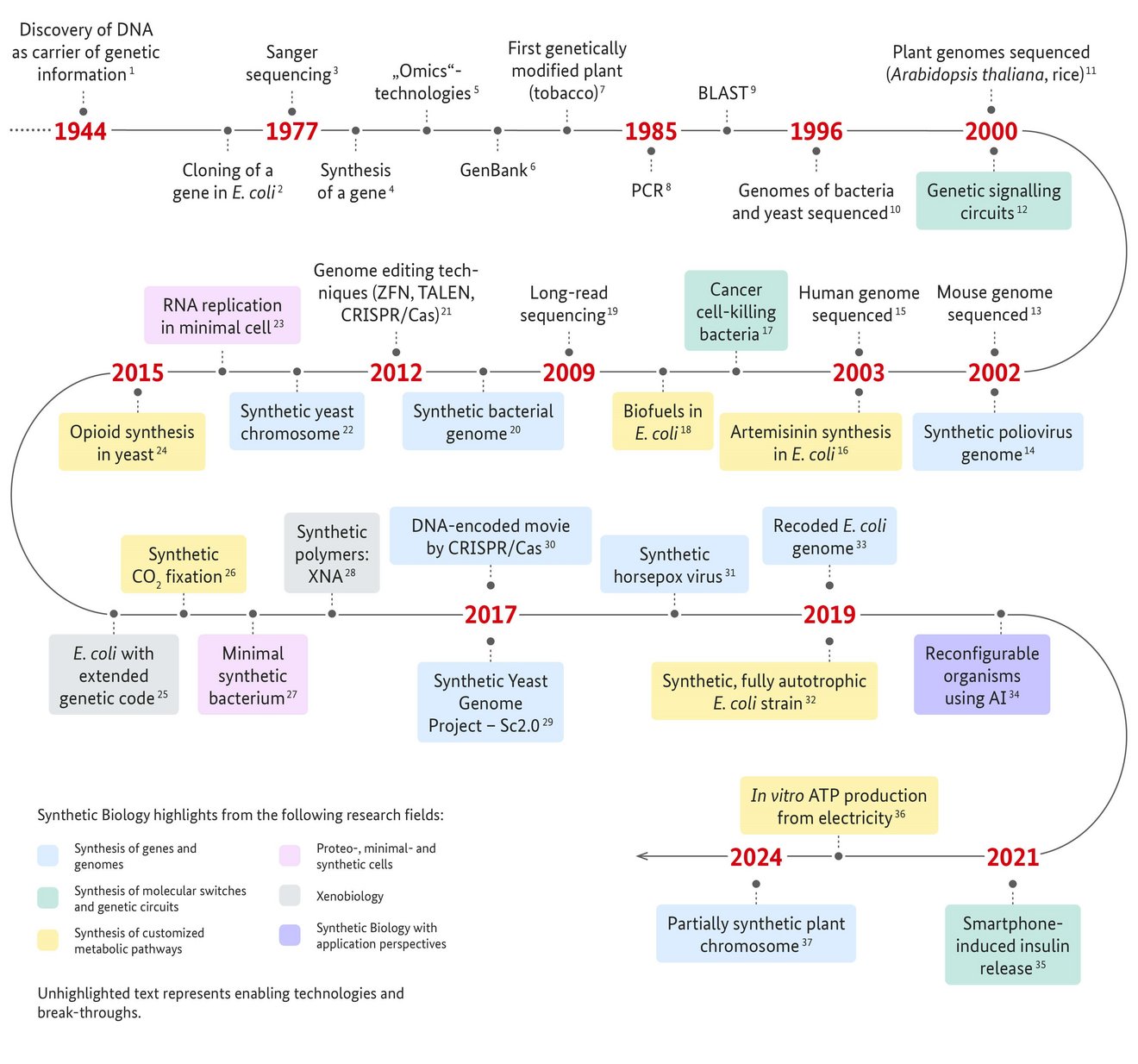
1 Avery et al., 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyriboenucleic acid fraction isolated from pneumococcus type III. J Exp Med 79 (2):137–58.
2 Chang & Cohen, 1974. Genome Construction Between Bacterial Species In Vitro: Replication and Expression of Staphylococcus Plasmid Genes in Escherichia coli. Proc Natl Acad Sci U S A 71(4):1030-4.
3 Sanger et al., 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74(12):5463-7.
4 Khorana et al., 1979. Total Synthesis of a Gene. Science 203(4381):614-25.
5 Review: Dai & Chen, 2022. Advances and Trends in Omics Technology Development. Front Med (Lausanne). 1(9): 911861.
6 https://www.ncbi.nlm.nih.gov/genbank/
Benson et al., 1993. GenBank. Nucleic Acids Res. 21(13):2963-5.
7 Bevan et al., 1983. A chimaeric antibiotic resistance gene as a selectable marker for plant cell transformation. Nature 304:184-7.
8 Patent US4683202A : Process for amplifying nucleic acid sequences. Filed Oct. 25, 1985; Date of Patent: Jul.28, 1987; Assignee: Cetus Corp, Inventor: Kary B. Mullis
Mullis et al., 1986. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 51 Pt 1:263-73.
9 Altschul et al., 1990. Basic local alignment search tool. J Mol Biol 215(3):403-10.
10 Fleischmann et al., 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269(5223):496-512.
Blattner et al., 1997. The complete genome sequence of Escherichia coli K-12. Science. 277(5331):1453-62.
Goffeau et al., 1996. Life with 6000 genes. Science 274(5287):546, 563-7.
11 The Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815.
Eckardt , 2000. Sequencing the rice genome. Plant Cell 12(11):2011-7.
12 Elowitz & Leibler, 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403(6767):335-8.
Review: MacDonald & Daens, 2016. Tools and applications in Synthetic Biology. Adv Drug Deliv Rev 105(Pt A):20-34.
13 Mouse Genome Sequencing Consortium, 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–62.
14 Cello et al., 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297(5583):1016-8.
15 International Human Genome Sequencing Consortium, 2004. Finishing the euchromatic sequence of the human genome. Nature 431(7011):931-45.
16 Martin et al., 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 21(7):796-802.
17 Anderson et al., 2006. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 355(4):619–27.
Review: Chien et al., 2017. Advances in Bacteria Cancer Therapies using Synthetic Biology. Curr Opin Syst Biol 5:1-8.
18 Kalscheuer et al., 2006. Escherichia coli engineered for fuel production. Microbiology (Reading). 2006 Sep;152(Pt 9):2529-36.
Review: Clomburg at al., 2010. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl Microbiol Biotechnol 86(2):419–34.
19 Eid et al., 2009. Real-time DNA sequencing from single polymerase molecules. Science 323(5910):133-8.
Clarke et al., 2009. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 4(4):265-70.
20 Gibson et al., 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329(5987):52-6.
21 Kim et al., 1996. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 93(3):1156–60.
Christian et al., 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 186(2):757-61.
Jinek et al., 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337(6096):816-21.
22 Annaluru et al., 2014. Total synthesis of a functional designer eukaryotic chromosome. Science 344(6179):55-8.
23 Caschera & Noireaux, 2014. Integration of biological parts toward the synthesis of a minimal cell. Curr Opin Chem Biol 22:85-91.
24 Galanie et al., 2015. Complete biosynthesis of opioids in yeast. Science 349(6252):1095-100.
25 Ostrov et al., 2016. Design, synthesis, and testing toward a 57-codon genome. Science 353(6301):819-22.
26 Schwander et al., 2016. A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354(6314):900-4.
27 Hutchison et al., 2016. Design and synthesis of a minimal bacterial genome. Science 351(6280):aad6253.
28 Acevedo -Rocha & Budisa, 2016. Xenomicrobiology: a roadmap for genetic code engineering. Microb Biotechnol 9(5):666-76.
29 Richardson et al., 2017. Design of a synthetic yeast genome. Science 355(6329):1040-44.
30 Shipman et al., 2017. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature 547(7663):345-9.
31 Noyce et al., 2018. Construction of an infectious horsepox virus vaccine from chemically synthesized DNA fragments. PLoS ONE 13(1): e0188453.
32 Gleizer et al., 2019. Conversion of Escherichia coli to Generate All Biomass Carbon from CO2. Cell 179(6):1255-63.e12.
33 Fredens et al., 2019. Total synthesis of Escherichia coli with a recoded genome. Nature 569(7757):514.8.
34 Kriegman et al., 2020. A scalable pipeline for designing reconfigurable organisms. Proc Natl Acad Sci U S A 117(4):1853-9.
35 Mansouri et al., 2021. Smartphone-Flashlight-Mediated Remote Control of Rapid Insulin Secretion Restores Glucose Homeostasis in Experimental Type-1 Diabetes. Small 17(35): e2101939.
36 Luo et al.,2023. ATP production from electricity with a new-tonature electrobiological module. Joule 7(8): 1745-58.
37 Chen et al., 2024. A designer synthetic chromosome fragment functions in moss. Nat Plants 10(2):228-39.